 This paper is dedicated to the memory of our friend and
colleague Vladimir M. Shulmeister, who was a key member on the project until
his untimely death on September 27, 1995.
This paper is dedicated to the memory of our friend and
colleague Vladimir M. Shulmeister, who was a key member on the project until
his untimely death on September 27, 1995.
Zhaolei Zhangac, Lishar Huangab, Vladimir M. Shulmeister, Young-In Chib, Kyeong Kyu Kimb, Li-Wei Hungc, Antony R. Croftsd, Edward A Berrya, and Sung-Hou Kimabc
E. O. Lawrence Berkeley National Laboratorya,
Department of Chemistryb, and the Graduate Group of Biophysicsc, University of California, Berkeley, CA, USA and Center for Biophysics and Computational Biology, University of Illinois at Urbana-Champaign, IL, USAd
 This paper is dedicated to the memory of our friend and
colleague Vladimir M. Shulmeister, who was a key member on the project until
his untimely death on September 27, 1995.
This paper is dedicated to the memory of our friend and
colleague Vladimir M. Shulmeister, who was a key member on the project until
his untimely death on September 27, 1995.
Introduction
Overall shape of the cytochrome bc1 complex dimer
Inhibitor binding sites
Arrangement of the protein domains in the intermembrane region
Structure of cytochrome c1
Structure and location of the Rieske ironsulfur protein
The extrinsic domain of the ironsulfur protein is loosely attached and functionally swapped between monomers.
Two conformations of the Rieske protein: Dynamic domain mediated electron transfer
Feasibility of electron transfer from the ironsulfur protein in the distal conformation to cytochrome c1.
Model for electron transfer through the high potential chain.
METHODS
References
Tables
Figure Legends
X-ray crystallographic analysis of multiple crystal forms of the cytochrome bc1 complex from three different species reveals two different locations for the extrinsic domain of the ironsulfur protein (ISP). One location is close to the putative quinol oxidation site, allowing reduction of the ISP by ubiquinol, and the other site is close enough to cytochrome c1 to allow rapid oxidation of the ISP by the cytochrome. However, neither location would allow both reactions to proceed at a kinetically competent rate. The reaction mechanism thus must involve a movement of the extrinsic domain of ISP, a mechanism not observed before for electron transport within redox protein complexes.
The energy conversion of the biosphere is achieved predominantly
through respiration and photosynthesis, and represents a flux several orders of
magnitude greater than all anthropogenic energy usage. The underlying
mechanism involves the coupling of electron transfer along a chain of redox or
photoredox enzymes to proton translocation across an organellar membrane in
which those redox components are embedded. This gives rise to an
electrochemical proton gradient across that membrane, which can be coupled to
energy-requiring processes including synthesis of ATP--a principle first
proposed by Peter Mitchell in his chemiosmotic hypothesis (1).
The central component of the electron transfer chain in mitochondria and in
many aerobic or photosynthetic bacteria is a complex of membrane proteins known
as the cytochrome bc1 complex, or ubiquinol:cytochrome c oxidoreductase (E.C.
1.10.2.2). This enzyme complex catalyzes electron transfer from ubiquinol to a
soluble cytochrome c, coupled to translocation of 2 H+ across the
inner mitochondrial membrane per quinol oxidized (2, 3, 4). The complex
isolated from beef heart consists of 11 different polypeptides (5, 6) with a
total molecular mass of 240 kDa (Table 1). There are four redox centers: two
hemes bH and bL of cytochrome b, one heme of cytochrome c1, and one ironsulfur
cluster of the Rieske protein. A mechanism accounting quantitatively for the
proton translocation coupled to electron transport by this enzyme is a version
of the "protonmotive Q cycle" of Peter Mitchell (3, 4). The mechanism also
explains the pattern of inhibition by the ubiquinone analogs, antimycin,
stigmatellin, UHDBT, myxothiazol, and MOA-stilbene, which react specifically at
one or the other of the two catalytic sites at which quinone is processed (3,
4).
Until recently only a low resolution structure for the complex from
Neurospora crassa has been available from electron microscopy of
two-dimensional crystals (7). In 1991 Yue et al. reported 3-D
crystallization of the bc1 complex from beef mitochondria in a tetragonal space
group (8). Two other groups independently reported crystallization of the beef
complex in different space groups in the following year (9, 10). Recently Yu
et al. (11) and Xia et al. (12) reported a partial structure of
the complex from the tetragonal crystals. In this structure, the extrinsic
domain of the Rieske protein was too disordered to be traced, and cytochrome c1
was only partially traced.
In the meantime we have obtained other crystal forms (13) from other species,
including one from chicken heart mitochondria which diffracts to 3.0 Å
resolution (Table 2). With these crystals we determined the structure of the
complex, including the functionally important Rieske ironsulfur protein and
cytochrome c1. We were also able to assign three additional subunits
(subunits 8, 10, and 11) that were not assigned before (12).
A comparison of our structures with and without various inhibitors reveals
that the ironsulfur cluster-containing extrinsic domain of the Rieske protein
assumes one of two conformations in the complexes. In one conformation the
ironsulfur cluster of the Rieske protein is close to its electron acceptor, the
heme of cytochrome c1, but far from the presumed binding site of its electron
donor, ubiquinol, in cytochrome b. In the other conformation the ironsulfur
cluster is closer to cytochrome b, and farther from cytochrome c1. This latter
conformation is similar to that found in the tetragonal beef crystals of Xia
et al. (12).
The binding sites for two Qo site inhibitors, stigmatellin and myxothiazol,
and the Qi site inhibitor, antimycin, have been located. The two Qo site
inhibitors bind in overlapping but not identical sites.
Taken together our two conformations for the ironsulfur protein and three
positions for ubiquinone analog inhibitors are compatible with all the
reactions proposed by the Q-cycle mechanism for electron transfer coupled to
proton translocation. However, no one structure alone would be competent. We
therefore propose that the reaction mechanism for electron transfer in the
cytochrome bc1 complex requires a dramatic conformational change involving
movement of the ironsulfur protein.
Overall shape of the cytochrome bc1 complex dimer
In all crystals of the complex from three sources (Table 1) the bc1
complex is present as a dimer (Figure. 1a), in which two monomers are related
by a two-fold axis running vertically in the plane of the paper. The protein
extends from the membrane 79 Å into the matrix space and 31 Å into
the inter-membrane region on either side of a transmembrane region 40 Å
thick, giving a total length of 150 Å perpendicular to the membrane. The
overall shape of the dimer is similar to that described for the beef complex by
Yu et al. (11) and Xia et al. (12), but considerably more protein
has been modeled in the inter-membrane domain. We have located subunits 1
through 8 and 10 in the electron density of the chicken crystal. We assign
subunit 10 to the transmembrane helix labeled N1 by Xia et al (12), based on
good correlation between side chains observed in our chicken map with the
sequence of this subunit from beef. Subunit 11 seems not to be present in our
preparation of the chicken enzyme, but is present in the beef and rabbit
enzymes. It probably corresponds to the transmembrane helix labeled N2 by Xia
et al., because this helix is present in the three crystal forms from the beef
and rabbit enzymes and not in the chicken crystals. However the resolution of
our mammal crystals is not high enough to confirm this by side chain
correlation with the sequence. Subunit 9 has not been assigned yet, and is
also missing from the structure of reference 12. This subunit is the
pre-sequence (14) of the Rieske protein, which gets cleaved off by a matrix
processing protease, so it is likely that at least its cleavage site is on the
matrix side of the membrane. We also see densities at a number of sites in the
transmembrane portion which we attribute to ubiquinone, detergents, and
phospholipids. These have not been modeled yet.
In the transmembrane domain the helices of the dimer fall into two clearly
separated, packed bundles. We arbitrarily divide the dimer so that one monomer
corresponds to one packed bundle of helices in the transmembrane region.
Inhibitor binding sites
The inhibitors antimycin, myxothiazol, and stigmatellin, when present
in stoichiometric excess during crystallization, resulted in electron density
increases that could be interpreted as due to the bound inhibitors. The
general positions of the antimycin, stigmatellin, and myxothiazol binding sites
are similar to those inferred from the figures in references 11 and 12.
Although the limited resolution does not allow detailed atomic model-building,
we have constructed speculative models consistent with the electron density.
These inhibitor binding sites, and especially the Qo site, are the target of
active drug design efforts to produce environmentally safe and effective plant
protection fungicides for agricultural use (15-17).
The antimycin site. Based on its mode of inhibition, antimycin is
thought to bind at the Qi site postulated in the Q-cycle mechanism, where
ubiquinone is reduced by electrons from cytochrome b with uptake of protons
from the matrix space (resulting in proton translocation when ubiquinol is
subsequently oxidized at the Qo site with proton release to the external
medium). The antimycin site (Figure 2a) was near the high potential heme of
cytochrome b, in a cavity surrounded by the heme, the transmembrane helices A,
D, and E, and the amphipathic surface helix a, with possible protonic
connection to the matrix phase via or around conserved histidine H202. The
close approach of the aromatic ring of the inhibitor to the heme was expected
from the effect of antimycin on the alpha absorption peak of the high potential
heme and the fluorescence quenching of antimycin when specifically bound at
this site (18). Residues F221, D21, and T194 are also close enough to contact
the inhibitor. One of the heme propionates is in van der Waals contact with
the inhibitor and curves around to form an ion pair with R101. The conformation
of this propionate, which differs from that depicted for the tetragonal beef
crystals (12), is the same in the absence of antimycin.
The stigmatellin site. The stigmatellin binding pocket (Figure 2b) is
formed by the C-terminal end of helix C, the helix cd1, the ef linker
(including the highly conserved -PEWY- sequence and the helix ef), and the
N-terminal end of helix F. Residues P271, F275, and M125 of cytochrome b and
H161 of the Rieske protein, which has moved from its position in the native
crystal (see below), are near the inhibitor. Residues 126-129 of helix C, and
140-147 of the linker cd, are also close by. In the native crystals Y279
passes through the region where we have modeled the stigmatellin head group,
but in the stigmatellin-complexed crystal Y279 has moved and is interacting
with R283 and with the Rieske backbone around C160.
The myxothiazol site. Myxothiazol (not shown) binds in roughly the
same place as stigmatellin, but displaced a little toward the center of the
membrane and toward the low potential b heme. It is also close to P271, but
where stigmatellin reaches outward from P271 toward the Rieske protein,
myxothiazol and MOA-stilbene reach toward Y132 and F129 in helix C, in the
vicinity of the low potential heme. This may be the site from which electron
transfer from the ubisemiquinone to the cytochrome bL heme occurs.
Arrangement of the protein domains in the intermembrane region
Figure 3 shows a slab including the extrinsic domains in the
inter-membrane region of the chicken complex. The two cytochrome c1 molecules
(purple) contact each other through loops which surround an empty area around
the two-fold axis. Subunit 8 (the "hinge protein for formation of the
cytochrome c1-c complex" of ref. 19, light blue) and the external ends of
subunits 7 (dark blue) and 10 (green) interact with cytochrome c1 on the side
away from the dimer interface. The hinge protein consists of a bent hairpin
held by two internal disulfide bonds.
Structure of cytochrome c1
Cytochrome c1 is one of the three redox-active proteins in the
cytochrome bc1 complex, but is incomplete in the structure of the beef complex
described by Xia et al. (12). This subunit is well ordered in our
chicken crystals and the entire polypeptide could be traced. Its extrinsic
domain forms a wedge-like structure containing the heme, with a C-terminal
transmembrane anchor adjacent to helix E of cytochrome b. Figure 4 compares
the backbone folding pattern of cytochrome c1 and mitochondrial cytochrome c, a
member of Ambler's Class I cytochromes c (20). Cytochrome c has five helical
segments, labeled
Mitochondrial cytochromes c have the pyrrole C corner of the heme exposed at
the "front" face, where electron transfer is believed to take place. This
corner is also exposed in our cytochrome c1 structure. The exposed C corner of
the heme is surrounded by three regions of the protein, consisting of residues
36-41 (corresponding to cytochrome c 13-18, "fingerprint" region), the
side-chain of Y95 and residues 104-106 (helix 2' , no corresponding residues in
cyt c), and 158-163 (containing the heme ligand M160 and corresponding to
cytochrome c 77-82).
Major differences between cytochromes c and c1 are the result of additions or
deletions in loop regions. For example, bovine cytochrome c1 has an N-terminal
extension of 24 residues before helix
There is a second exposure of the heme on the A-D edge: the long loop
corresponding to residues 41-58 in Tuna cytochrome c, which is present in
cytochromes c and c2, is absent in cytochrome c1. This results in exposure of
the heme propionates to the surface. As described below this edge is within
electron transfer distance of the ironsulfur cluster in some crystals,
suggesting this is the pathway for reduction by the ironsulfur protein.
Structure and location of the Rieske ironsulfur protein
Another of the three functionally important redox-active subunits of
the cytochrome bc1 complex, the Rieske ironsulfur protein, is missing in the
structure of the tetragonal beef crystal (12). Electron densities in the
region of the globular extrinsic domain of this protein in our crystals are
weaker than those in the rest of the structure, but clearly present and
recognizable (Figure 5). The backbone density is completely connected only
when contoured at 1
As predicted from hydropathy plots and molecular engineering results (24, 25),
the ironsulfur protein has a membrane-spanning helical segment near the
N-terminus. This was removed by proteolysis in preparing the soluble domain
for structure determination (23). However, the electron density in our map
(Figure 5a) continues where the model stops and connects to a transmembrane
helix. The transmembrane helix is well ordered.
The N-terminal 24 residues are on the matrix side, and interact with subunit
1. Residues 25 to 62 form a transmembrane helix, in close proximity with the
transmembrane helices of subunit 10 and cytochrome c1 (and, in the mammal
crystals, the putative subunit 11). The transmembrane helix is slightly
curved and highly slanted. It passes through the membrane at an angle of about
32deg. to the two-fold axis, assumed perpendicular to the membrane. This high
degree of tilt accounts for the length of the transmembrane helix (37
residues), which had led to suggestions of two transmembrane helices for the
Rieske protein (25).
Residues 60-66 are in close contact with both cytochrome b subunits in the
dimer, while residues 67-73 provide a flexible "tether" connecting the
extrinsic domain of the Rieske protein to its transmembrane helix. Figure 5b
shows a close-up of the ironsulfur cluster region of the Rieske protein. Two
histidine ligands, residues 141 and 161, are clearly evident as bulges in the
density at the tip of the protein. The iron atoms (oblong orange net) are not
individually resolved in this map.
The extrinsic domain of the ironsulfur protein is loosely attached and
functionally swapped between monomers.
Except for the transmembrane helix, only residues 141-143 of the ironsulfur
protein (one of the two loops which enclose the cluster) make contact with
cytochrome b in the native chicken crystals. This contact, depicted in Figure
5c, seems to involve interaction of L142 and G143 of the Rieske protein of one
monomer with T265 and L263 of cytochrome b of the other monomer.
The extrinsic domain of the ironsulfur protein has no contacts with the other
extrinsic domains within a monomer in the native chicken crystals (Figure 3).
But the ironsulfur cluster is close to the heme of cytochrome c1 of the
other monomer within the complex dimer. As described below this is
likely to be the pathway for electron transfer between the ironsulfur protein
and cytochrome c1. Taking monomers to be as defined above based on the
transmembrane region, the ironsulfur cluster is in a position to transfer
electrons with cytochromes b and c1 of the other monomer.
The small number of contacts with the rest of the dimer probably accounts for
the poor order of the Rieske extrinsic domain, and suggests that the domain is
mobile. This mobility is somewhat restricted in one monomer of the chicken
crystals and in the beef and rabbit hexagonal crystals by inter-dimeric crystal
contacts involving the extrinsic domain of the Rieske protein. Xia and
coworkers (12) also concluded that the ironsulfur protein is highly mobile.
They suggested that mobility may be required for function, based on poor order
of this domain in their crystals and the large distance between the cluster and
the heme of cytochrome c1.
Two conformations of the Rieske protein: Dynamic domain mediated electron
transfer
While the distances between the six heme iron peaks of the dimer were
the same within experimental error in all four crystal forms, the distance from
the iron-sulfur cluster to any heme varied by up to 5 Å in the different
native crystals. Based on published distances between iron centers, the
ironsulfur cluster in the tetragonal beef crystals (11, 12) is in a markedly
different position than in any of our native crystals (Table 3). But when the
chicken cytochrome bc1 complex was treated with a saturating amount of the
inhibitor stigmatellin before crystallization, we found the extrinsic domain of
the ironsulfur protein at a location different from that in native crystals,
and the ironsulfur cluster at a location similar to that reported for the
tetragonal beef crystals (12). This movement can be simply and dramatically
demonstrated using Bivoet difference maps constructed from diffraction data
collected with X-ray wavelength near the iron absorption edge. Due to anomalous
scattering by iron, the peaks in such maps indicate positions of irons in the
complex: 3 heme irons of the cytochromes and the ironsulfur cluster of the
Rieske protein. Such maps are shown in Figure 6, in the absence (a) or
presence (b) of stigmatellin. The peak labeled Fe2S2 increases in intensity
and moves closer to the hemes of cytochrome b in the presence of stigmatellin.
We call this the proximal conformation of the Rieske protein, and the
conformation in our native crystals the distal conformation. The relative
position of the ironsulfur cluster in the chicken crystals with stigmatellin is
16 Å from the position in the native chicken crystals, and 20 Å
from that in the beef hexagonal crystals.
A stereo view of the two conformations of the Rieske protein in context of the
entire bc1 complex dimer and in isolation with cytochrome b and the heme of
cytochrome c1 are shown in Figures 1b and c. The two locations of the
extrinsic domain of the Rieske protein are related by a rotation of 57deg.
about an axis passing near residues 93 and 182 of the protein, perpendicular to
the plane of the picture in Figure 1c. The transmembrane helix and matrix-side
portion are unchanged in the presence of stigmatellin. The coil consisting of
residues 68-73 is stretched out in the presence of stigmatellin, allowing this
end of the soluble domain to move farther from the membrane as the Fe2S2
cluster on the other end moves closer. In a crystal with both stigmatellin and
antimycin bound, the ironsulfur position was essentially the same as that with
only stigmatellin. In crystals with only antimycin or myxothiazol bound the
ironsulfur position was similar to that in the crystals with no inhibitors.
In the proximal conformation the ironsulfur cluster of the Rieske protein is
in H-bond distance of the occupant of the Qo site - stigmatellin in our
crystals (Figure 2b) but by inference the electron donor ubiquinol in
vivo - and in the distal conformation the ironsulfur cluster is close to
its electron acceptor, the heme of cytochrome c1. This suggests that the
reaction mechanism for electron transfer in the cytochrome bc1 complex requires
this dramatic conformational change involving movement of the extrinsic domain
of the ironsulfur protein.
Feasibility of electron transfer from the ironsulfur protein in the distal
conformation to cytochrome c1. In the native chicken crystals the second
loop of the Rieske protein enclosing the cluster (residues around H161) faces
toward cytochrome c1, approaching the heme propionates and residues 106 and 145
of cytochrome c1. There is, however, no electron density contact with
cytochrome c1 in the chicken crystals when contoured at the 1.0
As shown in Table 3, the Rieske protein is closer to cytochrome c1 in our two
beef crystals. In these crystals there is electron density contact at the 2
Model for electron transfer through the high potential chain.
Figure 7 shows a ribbon diagram of the extrinsic domains of the Rieske
ironsulfur protein and cytochrome c1, as well as cytochrome c bound to
cytochrome c1 at a hypothetical site and orientation. The position of the
ironsulfur protein is that from the chicken crystals (in the beef crystals it
is about 4 Å closer to cytochrome c1). This diagram illustrates the
possibility of electron transfer into cytochrome c1 via the D propionate and
out of cytochrome c1 via the C corner of the heme to cytochrome c. The
distance between the two cytochromes is 10.2 Å, measured between atoms
C2C of each heme (the closest approach of the
METHODS
Purification and crystallization
The cytochrome bc1 complex was purified from different vertebrate heart
tissues essentially as described for the potato complex (28). Mitochondria
were prepared by the method of Smith (29) and solubilized using the detergent
dodecyl maltoside. The complex was isolated from the extract by chromatography
on DEAE Sepharose CL6B and further purified by size exclusion chromatography on
Sepharose CL6B (13). The protein was concentrated to around 200 uM by
ultrafiltration through an Amicon YM-100 membrane, precrystallized by mixing
with 100 mM KMES pH 6.5 and 10% PEG-4000, and redissolved in 20 mM K-MOPS 7.5,
20 g/L n-octyl-
To co-crystallize the bc1 complex with the high-affinity inhibitors antimycin,
myxothiazol, or stigmatellin, the inhibitor was added from an ethanolic
solution (the final ethanol concentration was below 1% v/v) in a 1.5-2.0-fold
molar ratio to the pooled fractions from the final column at a protein
concentration of 5-10
Cryogenic data collection and reduction
After crystallization was complete (five to thirty days after setup),
20 ul of cryoprotectant containing 10 mM K-MES pH 6.7, 10 mM
n-octyl-
Structure determination
The chicken crystals were phased by isomorphous replacement and the
resulting electron density was used to phase the other crystal forms by
molecular replacement. Heavy atom derivatives were first analyzed using
XtalView (31). The RAVE package (32) was used for molecular averaging, map
skewing, and rotation-translation operator improvement. The CCP4 package (33)
was used for final heavy atom refinement and phase calculation (program
MLPHARE), and for finding molecular replacement solutions (programs ALMN and
TFFC). The phases were improved and extended to the resolution limit of the
data by multi-crystal and noncrystallographic symmetry averaging. During the
phase improvement and extension process, correlation coefficients between the
calculated electron density map of the Rieske protein and our experimental
electron density, which were monitored as a measure of the improvement of the
maps, increased to 80-85% in the different data sets. The coefficient between
subunits 1 and 2 increased to 40-48%.
Model building was done with the program O (34), and the structures were
illustrated using this program or Molscript (35) and Raster3D (36).
All subunits of the bc1 complexes of vertebrates are expressed in the
cytoplasm, except cytochrome b. Cytochrome b, which is expressed in
mitochondria, has sequence identity of 74% between the chicken and beef
proteins. Since amino acid sequences of chicken subunits of the complex have
not been reported, we have used the sequences of the beef proteins for the
remainder in our model building. Cytochrome c, a cytoplasmically expressed
mitochondrial protein, has 89.5% identity between chicken and beef. Myosin
light chain two of chicken is 91% identical to that from human or mouse.
Location of iron centers from anomalous data
Anomalous data at wavelength near the iron K absorption edge (7131 eV)
were collected for native and stigmatellin-containing crystals, and Bivoet
difference maps with coefficients of (F+ - F-) and
improved experimental phases retarded by 90deg. were made to locate the iron
centers.
Electron density map calculations
The electron density maps were calculated using coefficients of
(2Fo-Fc) e-i
Correspondence and requests for materials to: Dr. Edward A. Berry and Prof.
Sung-Hou Kim, Calvin Laboratory # 5230, University of California, Berkeley,
Berkeley, CA 94720-5230, U.S.A. Atomic coordinates of chicken cytochrome bc1
complex have been deposited in the Brookhaven Protein Database for release in
May 1998 (entry XXXX for the native chicken structure and YYYY for the
stigmatellin+antimycin inhibited chicken structure).
1. Mitchell, P. Coupling of phosphorylation to electron and proton transfer by
a chemiosmotic type of mechanism. Nature (London) 191, 144-148
(1961).
2. Hatefi, Y., Galante, Y. M., Stiggall, D. L. & Ragan, C. I. Proteins,
polypeptides, prosthetic groups, and enzymic properties of complexes I, II,
III, IV, and V of the mitochondrial oxidative phosphorylation system.
Methods Enzymol. 56, 577-602 (1979).
2. Hinkle, P. C., Kumar, M. A., Resetar, A. & Harris, D. L. Mechanistic
Stoichiometry of Mitochondrial Oxidative Phosphorylation. Biochemistry
30, 3576-3582 (1991).
3. Mitchell, P. Possible molecular mechanisms of the protonmotive function of
cytochrome systems. J. Theor. Biol. 62, 327-367 (1976).
4. Crofts, A. R. The mechanism of Ubiquinol:cytochrome c Oxidoreductases of
Mitochondria and of Rhodopseudomonas sphaeroides. in The Enzymes of
Biological Membranes, Vol. 4 (ed. Martonosi, A. N.) 347-382 (Plenum
Publishing Corporation, New York, 1985).
5. Schägger H., Link, Th. A., Engel, W. D. & von Jagow, G. Isolation
of the Eleven Protein Subunits of the bc1 complex from beef heart. Methods
in Enzymology 126, 224-237 (1986).
6. Schaegger, H., Brandt, U., Gencic, S. and von Jagow, G.,
Ubiquinol-cytochrome-c reductase from human and bovine mitochondria
Methods in Enzymol. 260, 82-96. (1995)
7. Weiss, H. & Leonard, K. Structure and Function of Mitochondrial
Ubiquinol:Cytochrome c Reductase and NADH:Ubiquinone Reductase. Chemica
Scripta 27B, 73-81 (1987).
8. Yue, W. H., Zou, Y. P., Yu, L. & Yu, C. A. Crystallization of
mitochondrial ubiquinol-cytochrome c reductase. Biochemistry 30,
2303-2306 (1991).
9. Kubota T., Kawamoto M., Fukuyama K., Shinzawa-Itoh, K., Yoshikawa, S. &
Matsubara, H. Crystallization and preliminary X-ray crystallographic studies of
bovine heart mitochondrial cytochrome bc1 complex. J. Mol. Biol. 221,
379-382 (1991).
10. Berry, E. A., Huang, L.-S., Earnest, T. N. & Jap, B. K. X-ray
Diffraction by Crystals of Beef Heart Ubiquinol:Cytochrome c Oxidoreductase.
J. Mol. Biol. 224, 1161-1166 (1992).
11. Yu, C.-A., Xia, J.-Z., Kachurin, A. M., Yu, L., Xia, D., Kim, H. &
Deisenhofer, J. Crystallization and preliminary structure of beef heart
mitochondrial cytochrome-bc1 complex. Biochim. Biophys. Acta
1275, 47-53 (1996).
12. Xia, D., Yu, C.-A., Kim, H., Jia-Zhi Xia, J.-Z., Anatoly, M., Kachurin, A.
M., Zhang, L., Yu, L. & Deisenhofer, J. Crystal Structure of the Cytochrome
bc1 Complex from Bovine Heart Mitochondria. Science 277, 60-66
(1997).
13. Berry, E. A., Huang, L.-S., Shulmeister, V. M. & Kim, S.-H. A New Form
of Crystal of Bovine Heart Ubiquinol:Cytochrome c Oxidoreductase--Determination
of Space Group and Unit Cell Parameters. Acta Cryst. D51, 235-239
(1995).
14. Brandt, U., Yu, L., Yu, C. A. & Trumpower, B. L. The mitochondrial
targeting presequence of the Rieske iron-sulfur protein is processed in a
single step after insertion into the cytochrome bc1 complex in mammals and
retained as a subunit in the complex. J. Biol. Chem. 268,
8387-8390 (1993).
15. Beautement, K., Clough, J. M., Defraine, P. J. & Godfrey, C. R. A.
Fungicidal Beta- Methoxyacrylates--From Natural Products To Novel Synthetic
Agricultural Fungicides. Pesticide Science 31, 499-519 (1991).
16. Clough, J. M. & Godfrey, C. R. A. Growing Hopes. Chemistry In
Britain 31, 466-469 (1995).
17. Sauter, H., Ammermann, E. & Roehl, F. Strobilurins--From natural
products to a new class of fungicides. in Crop Protection Agents from Nature
(ed. Copping, L. G.) 50-81 (The Royal Society of Chemistry, Thomas Graham
House, Cambridge, UK, 1996).
18. Slater, E. C. The mechanism of action of the respiratory inhibitor,
antimycin. Biochim. Biophys. Acta 301, 129-154 (1973).
19. Kim, C.H. and King, T.E. A Mitochondrial Protein Essential for the
Formation of the Cytochrome c1-c complex. Isolation, purification, and
properties. J. Biol. Chem 258, 13543-51 (1983)
20. Ambler, R.P. The Structure and Classification of Cytochromes c in From
Cyclotrons To Cytochromes (eds. N.O. Kaplan & A. Robinson), pp. 263-280
(Academic Press, New York, 1982), and Sequence Variability in Bacterial
Cytochromes c, Biochim. Biophys. Acta 1058, 42-47, (1991)
21. Stonehuerner, J., O'Brien, P., Geren, L., Millett, F., Steidl, J., Yu, L.,
& Yu, C.-A. Identification of the Binding Site on Cytochrome c1 for
Cytochrome c. J. Biol. Chem. 260, 5392-5398 (1985).
22. Broger, C., Salardi, S. & Azzi, A. Interaction between Isolated
Cytochrome c1 and cytochrome c. Eur. J. Biochim. 131, 349-352
(1983).
23. Iwata, S., Saynovits, M., Link, Th. A. & Michel, H. Structure of a
water soluble fragment of the 'Rieske' iron-sulfur protein of the bovine heart
mitochondrial cytochrome bc1 complex determined by MAD phasing at 1.5Å
resolution. Structure 4, 567-579 (1996).
24. Van Doren, S. R., Yun, C.-H., Crofts, A. R. & Gennis, R. Assembly of
the Rieske iron- sulfur subunit of the cytochrome bc1 complex in Escherichia
coli and Rhodobacter sphaeroides membranes independent of the
cytochrome b and c1 subunits. Biochemistry 32, 628-636 (1993).
25. Link, T. A., Schägger, H. & von Jagow, G. Structural analysis of
the bc1 complex from beef heart mitochondria by the sidedness hydropathy plot
and by comparison with other bc complexes. in Cytochrome Systems: Molecular
Biology and Bioenergetics (eds. Papa, S., Chance, B. & Ernster, L.)
289-301 (Plenum Press, New York, 1987).
26. Moser, C.C., Page, C.C., Farid, R. and Dutton, P.L. (1995) Biological
electron transfer. J. Bioenergetics Biomembranes 27, 263-274.
26a. Ding, H., Moser, C.C., Robertson, D.E., Tokito, M.K., Daldal, F., and
Dutton, P.L. Ubiquinone pair in the Qo site Central to the Primary Energy
Conserving Reactions of Cytochrome bc1 complex. Biochemistry 34,
15979-96 (1995)
27. Crofts, A.R. and Wang, Z. Photosynthesis Research 22, 69-87
(1989)
28. Berry, E. A, Huang, L.-S. & DeRose, V. Ubiquinol-Cytochrome c
Oxidoreductase of Higher Plants. Isolation and Characterization of the bc1
Complex from Potato Tuber Mitochondria. J. Biol. Chem. 266,
9064-9077 (1991).
29. Smith, A. L. Preparation, properties, and conditions for assay of
mitochondria: slaughterhouse material, small scale. Meth. Enzymol.
10, 81-86 (1967).
30. Otwinowski, Z. Oscillation Data Reduction Program. in Proceedings of the
CCP4 Study Weekend: "Data Collection and Processing" (eds. Sawyer, L.,
Isaacs, N. & Bailey, S.) 56- 62 (SERC Daresbury Laboratory, England,
1993).
31. McRee, D. E. A visual protein crystallographic software system for
X11/XView. J. Molecular Graphics 10, 44-46 (1992).
32. Jones, T. A. Proc. CCP4 Study Weekend, Molecular Replacement (eds.
Dodson, E. J., Glover, S., & Wolf, W.) 91-105 (SERC Daresbury Laboratory,
UK, 1992).
33. CCP4 The SERC (UK) Collaborative Computing Project No. 4. The CCP4 Suite:
Programs for Protein Crystallography. Acta Cryst. D50, 760-763
(1995).
34. Jones, T. A., Zou, J. Y., Cowan, S. W. & Kjeldgaard, M. Improved
Methods For Building Protein Models In Electron Density Maps And The Location
Of Errors In These Models. Acta Crystallographica A47, 110-119
(1991).
35. Kraulis, P. J. MOLSCRIPT: a program to produce both detailed and schematic
plots of protein structures. Journal of Applied Crystallography 24,
946-950 (1991).
36. Merritt, E. A. & Murphy, M. E. P. Raster3D Version 2.0-A Program For
Photorealistic Molecular Graphics. Acta Crystallographica D50,
869-873 (1994).
37. Rayment, I. Molecular replacement method at low resolution: optimum
strategy and intrinsic limitations as determined by calculations on icosahedral
virus models. Acta Cryst. A39, 102-116 (1983).
Acknowledgments: We thank Thomas Link and his co-workers for providing
coordinates for the soluble domain of the Rieske ironsulfur protein before
their release from the Protein Data Bank, Henry Bellamy for performing the XAFS
scan and for advice on MAD data collection, Sangjin Hong for preparing
coordinate files for the inhibitors, and Liang Tong and Dave Schuller for
advice on molecular averaging. This investigation was supported by NIH (grants
to EAB and ARC) and by the Office of Biosciences and Environmental Research,
U.S. Department of Energy (grant to Sung-Hou Kim). The work was partially done
at SSRL which is operated by the Department of Energy, Division of
Chemical/Material Sciences. The SSRL Biotechnology Program is supported by the
National Institutes of Health Biomedical Resource Technology Program, Division
of Research Resources.
Tables
Table 1. Subunits of bovine heart cytochrome bc1 complex
Table 4. Structure determination statistics.
(a) Diffraction data for chicken crystals. Each line corresponds to one
dataset collected from a single native or derivatized crystal of chicken
cytochrome bc1.
(b) Isomorphous phase determination
Figure 1. Stereoview ribbon diagrams of the bc1 complex. (a)
The native chicken bc1 dimer. The molecular two-fold axis runs vertically
between the two monomers. The key for the color coding of each subunit is
given in the inset. The presumed membrane bilayer is represented by a gray
band. (b) Two conformations of the Rieske protein in one monomer shown in
context of the entire dimer. One conformation found in our native chicken
crystal (yellow) is superimposed on the other conformation (blue) from crystals
grown in the presence of stigmatellin (green). The hemes (red) of cytochrome
c1 and cytochrome b as well as two positions of the ironsulfur clusters of the
Rieske proteins are shown in orange and green. (c) Isolated close-up of the two
conformations of the Rieske protein in contact with cytochrome b with
associated hemes (red), stigmatellin (green) and antimycin (purple). The
isolated heme of cytochrome c1 (red) is also shown. The rotation axis relating
the two positions of the Rieske protein is indicated (white cross).
Figure 2. Inhibitor binding sites. The electron density maps are from
crystals containing the inhibitors. (a) antimycin binding site, electron
density map contoured at 0.7
Figure 3. Structure of the inter-membrane (external surface) domains of the
chicken bc1 complex. Viewed from within the membrane, with
the transmembrane helices truncated at approximately the membrane surface. Ball
and stick models represent the heme of cyt c1, the Rieske ironsulfur cluster,
and the disulfide cysteines of subunit 8.
Figure 4. The structure of cytochrome c1 compared to cytochrome c.
Top: ribbon diagram of mitochondrial cytochrome c with the open corner of C
pyrrole of the heme facing the viewer, and the heme propionates directed
downward. Bottom: our current structure of cytochrome c1, rotated to put the
common features between the two cytochromes in the same orientation.
Corresponding segments of each cytochrome are drawn with the same color.
Helices labeled
Figure 5. The Rieske ironsulfur protein. The electron density maps are
from improved experimental phases. The atomic model of the soluble domain of
the ironsulfur protein is from the coordinates of the protein database entry
1RIE (28), positioned as described in the text. (a) a slab through the protein
including the cluster and the connection to the transmembrane helix (b) a
closeup of the electron density around the ironsulfur cluster of the Rieske
protein. The maps are contoured at 1
Figure 6. Relative positions of the redox centers in the two different
conformations of the bc1 complex dimer. a. iron centers revealed by Bivoet
difference maps near the iron edge. b. Schematic drawing representing the
cofactors. Left: from a native crystal, with the ironsulfur cluster of the
Rieske protein in distal position from the low-potential heme of cytochrome b.
Right: from a crystal with bound stigmatellin, with the cluster in proximal
position.
Figure 7. Electron pathway through cytochrome c1 in a hypothetical complex
of the bc1 complex with cytochrome c. The ribbon diagram shows the
backbones of cytochrome c1, cytochrome c (both with the same color scheme as in
Figure 4), and the Rieske protein (yellow). The hemes, the ironsulfur cluster,
and surrounding residues are drawn as ball-and-stick models. The balls
representing the ironsulfur cluster (red & green) are enlarged for
visibility. The position of the Rieske protein relative to cyt. c1 is from the beef hexagonal crystals.
 1-
1-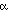 5. Three helices (
5. Three helices (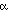 1,
1,
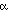 3, and
3, and 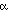 5), which are conserved in Class I cytochromes in
general, are present in cytochrome c1 and occupy the same positions relative to
each other and to the heme. They are colored alike and labeled on both
molecules in Figure 4. Conserved aromatic residues involved in interaction
between
5), which are conserved in Class I cytochromes in
general, are present in cytochrome c1 and occupy the same positions relative to
each other and to the heme. They are colored alike and labeled on both
molecules in Figure 4. Conserved aromatic residues involved in interaction
between 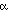 1 and
1 and 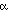 5 (F10 and Y97 in mitochondrial cytochrome) are
present as Y33 and F189, respectively. The tripeptide PNL starting at residue
30 is conserved in mitochondrial cytochromes c. The proline carbonyl accepts a
hydrogen bond from Nd of the histidine heme ligand and the leucine
provides hydrophobic environment for the heme ring. This aligns with the
tripeptide PDL starting at residue 111 of cytochrome c1. It is conserved in all
cytochromes c1 except that of Rhodobacter sphaeroides, which, barring a
sequencing error, has ADL. These similarities justify inclusion of cytochrome
c1 in the family of class I cytochromes.
5 (F10 and Y97 in mitochondrial cytochrome) are
present as Y33 and F189, respectively. The tripeptide PNL starting at residue
30 is conserved in mitochondrial cytochromes c. The proline carbonyl accepts a
hydrogen bond from Nd of the histidine heme ligand and the leucine
provides hydrophobic environment for the heme ring. This aligns with the
tripeptide PDL starting at residue 111 of cytochrome c1. It is conserved in all
cytochromes c1 except that of Rhodobacter sphaeroides, which, barring a
sequencing error, has ADL. These similarities justify inclusion of cytochrome
c1 in the family of class I cytochromes. 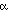 1, compared to 1 residue in
bovine cytochrome c. In cytochrome c1 this region interacts with subunit 8,
the hinge protein. After helix
1, compared to 1 residue in
bovine cytochrome c. In cytochrome c1 this region interacts with subunit 8,
the hinge protein. After helix 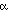 1 and the "fingerprint" CXYCH
heme-binding stretch, which are similar in the two cytochromes, cytochrome c1
has a long insertion (residues 52-109) between residues 18 and 29 of cytochrome
c. This expanded loop includes a region implicated for cytochrome c binding
(21) and the dimer contact with cytochrome c in the other monomer seen in
Figure 3. Another insertion is found between the methionine heme ligand and
helix 5, the six residues 81-86 in cytochrome c correspond to 18 (161-178) in
the c1 cytochromes. This region has also been implicated in cytochrome c
binding (22). The end of helix
1 and the "fingerprint" CXYCH
heme-binding stretch, which are similar in the two cytochromes, cytochrome c1
has a long insertion (residues 52-109) between residues 18 and 29 of cytochrome
c. This expanded loop includes a region implicated for cytochrome c binding
(21) and the dimer contact with cytochrome c in the other monomer seen in
Figure 3. Another insertion is found between the methionine heme ligand and
helix 5, the six residues 81-86 in cytochrome c correspond to 18 (161-178) in
the c1 cytochromes. This region has also been implicated in cytochrome c
binding (22). The end of helix 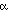 5 is the C-terminus of cytochrome c
but the transmembrane helix a6' continues after in cytochrome c1.
5 is the C-terminus of cytochrome c
but the transmembrane helix a6' continues after in cytochrome c1. 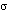 level or lower, whereas the cytochrome b backbone
in the transmembrane helices was continuous even when contoured at 3
level or lower, whereas the cytochrome b backbone
in the transmembrane helices was continuous even when contoured at 3 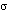 .
However, the density was good enough to unambiguously locate the known
structure of the soluble domain of the Rieske protein (23).
.
However, the density was good enough to unambiguously locate the known
structure of the soluble domain of the Rieske protein (23).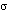 level.
level. 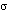 level between the Rieske protein around C160 (which forms a disulfide
bond holding the cluster-binding loops together) and cytochrome c1 around G107
(between helix
level between the Rieske protein around C160 (which forms a disulfide
bond holding the cluster-binding loops together) and cytochrome c1 around G107
(between helix 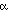 2' and the heme-bracing proline P111). This very
likely represents the configuration of the ironsulfur protein during electron
transfer to the cytochrome. H161 of the Rieske protein, which provides one of
the ligands to the Fe2S2 cluster, is 4.0 Å from an oxygen atom of heme
propionate D and 8.2 Å from the edge of the heme
2' and the heme-bracing proline P111). This very
likely represents the configuration of the ironsulfur protein during electron
transfer to the cytochrome. H161 of the Rieske protein, which provides one of
the ligands to the Fe2S2 cluster, is 4.0 Å from an oxygen atom of heme
propionate D and 8.2 Å from the edge of the heme  -bonded system at
the C3D atom. From this distance (8.2 Å) we can calculate an approximate
rate of electron transfer from the ironsulfur protein to cytochrome c1 of 4.8 -
80. x 106 s-1, assuming nonadiabatic electron tunneling
with a reorganization energy
-bonded system at
the C3D atom. From this distance (8.2 Å) we can calculate an approximate
rate of electron transfer from the ironsulfur protein to cytochrome c1 of 4.8 -
80. x 106 s-1, assuming nonadiabatic electron tunneling
with a reorganization energy 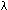 of 0.7 to 1.0 electron volts and
of 0.7 to 1.0 electron volts and  G near
zero (26). This is significantly faster than measured rates for this reaction
(27) so if the protein spends a small fraction of time in this conformation it
could account for the rate. In the native chicken crystals this distance is
14.4 Å, which would give a rate of 1.8 - 15. x
103s-1) with the same assumptions. In the crystal with
stigmatellin, or the tetragonal beef crystals (12) the shortest distance from
the cluster or its ligands to the heme tetrapyrrole ring is 27 Å, giving
with the same assumptions a rate of 10-4 s-1 and making
it very unlikely that the enzyme could function in this single conformation.
G near
zero (26). This is significantly faster than measured rates for this reaction
(27) so if the protein spends a small fraction of time in this conformation it
could account for the rate. In the native chicken crystals this distance is
14.4 Å, which would give a rate of 1.8 - 15. x
103s-1) with the same assumptions. In the crystal with
stigmatellin, or the tetragonal beef crystals (12) the shortest distance from
the cluster or its ligands to the heme tetrapyrrole ring is 27 Å, giving
with the same assumptions a rate of 10-4 s-1 and making
it very unlikely that the enzyme could function in this single conformation. -bonded systems). Assuming
-bonded systems). Assuming
 G near zero and reorganization energy
G near zero and reorganization energy 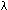 in the range 0.7 to
1.0 gives electron transfer rates in the range of 0.6-5.1 x 106
s-1.
in the range 0.7 to
1.0 gives electron transfer rates in the range of 0.6-5.1 x 106
s-1.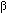 -D-glucopyranoside, and 100 mM NaCl. Aliquots (5-20
-D-glucopyranoside, and 100 mM NaCl. Aliquots (5-20 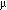 l)
were mixed with an equal volume of precipitant containing 20 mM KMES pH 6.7, 75
mM NaCl, 10% glycerol, and 6% PEG 4000; and subjected to vapor diffusion
against 30% glycerol.
l)
were mixed with an equal volume of precipitant containing 20 mM KMES pH 6.7, 75
mM NaCl, 10% glycerol, and 6% PEG 4000; and subjected to vapor diffusion
against 30% glycerol. 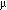 M before concentrating and precrystallizing as above.
M before concentrating and precrystallizing as above.
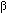 -D-glucopyranoside, 25% glycerol, and 10% PEG 4000 was added to
the solution containing the crystals from chicken complex, and the reservoir
was changed to 35% glycerol for further concentration of glycerol and PEG
without increasing ionic strength. After this equilibration or in some cases
after further soaking in cryoprotectant consisting of 30% glycerol, K-MES, and
n-octyl-
-D-glucopyranoside, 25% glycerol, and 10% PEG 4000 was added to
the solution containing the crystals from chicken complex, and the reservoir
was changed to 35% glycerol for further concentration of glycerol and PEG
without increasing ionic strength. After this equilibration or in some cases
after further soaking in cryoprotectant consisting of 30% glycerol, K-MES, and
n-octyl-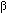 -D-glucopyranoside, crystals were frozen in liquid ethane or
nitrogen, or in the cryogenic stream, and data were collected at 70deg.-100deg.
K. A suitable procedure for flash-freezing the beef and rabbit crystals has
not yet been developed. Diffraction data were processed by the programs DENZO
and SCALEPACK (30).
-D-glucopyranoside, crystals were frozen in liquid ethane or
nitrogen, or in the cryogenic stream, and data were collected at 70deg.-100deg.
K. A suitable procedure for flash-freezing the beef and rabbit crystals has
not yet been developed. Diffraction data were processed by the programs DENZO
and SCALEPACK (30). 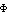 c, where the Fo values are from the
experimentally determined intensities but the Fc and
c, where the Fo values are from the
experimentally determined intensities but the Fc and 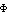 c are calculated
from the previous map after multiple crystal averaging. In the case of
unobserved reflections, Fo was replaced by Fc as recommended (37), resulting in
coefficients of Fc e-i
c are calculated
from the previous map after multiple crystal averaging. In the case of
unobserved reflections, Fo was replaced by Fc as recommended (37), resulting in
coefficients of Fc e-i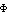 c for those terms. This "fill-in"
procedure was used both during averaging and, unless otherwise noted, in making
the final maps used in the figures.
c for those terms. This "fill-in"
procedure was used both during averaging and, unless otherwise noted, in making
the final maps used in the figures.
References
Subunit Residues MW
1 Core 1 446 49132
2 Core 2 439 46471
3 Cytochrome b 379 42592
4 Cytochrome c1 241 27288
5 Rieske Fe-S 196 21611
6 13.4 kDa 110 13347
7 "Q-binding" 81 9590
8 c1 "hinge" 78 9170
9 Fe-S preseq. 78 7956
10 c1-assoc. 62 7198
11 6.4 kDa 56 6363
Apo-bc1 complex 2166 240718
Fe2S2 76
Heme c1 616
Heme bH 616
Heme bL 616
Prosthetic groups 2024
Holo-bc1 complex 242,742 Da
Table
2. Parameters of crystal forms used in the structure determination
#AU Mono
Protein Space Group Unit Cell cell AU Å3/Da Dmax1
Beef bc1 a b c
Table
3. Distances between the Rieske ironsulfur cluster and cytochromes c1 and bL
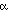
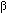
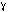 Hex bipyramid P6522 217 217 378 90deg. 12 1 5.20 3.8
90deg. 120deg.
Monoclinic2 P21 118 178 200 90deg. 2 2 4.16 3.7
106deg. 90deg.
Rabbit bc1
Hex bipyramid P6522 212 212 354 90deg. 12 1 4.60 3.5
90deg. 120deg.
Chicken bc1
Orthorhombic P212121 176 184 242 90deg. 4 2 4.04 3.0
90deg. 90deg.
frozen- P212121 169 181 239 90deg. 4 2 3.76 2.9
90deg. 90deg.
1. AU = asymmetric unit. Mono = monomer. Dmax = d-spacing of recorded indexable
reflection with highest resolution.
2. The monoclinic beef crystals were mistakenly reported as centered orthorhombic
(C2221) in ref. 13
Hex bipyramid P6522 217 217 378 90deg. 12 1 5.20 3.8
90deg. 120deg.
Monoclinic2 P21 118 178 200 90deg. 2 2 4.16 3.7
106deg. 90deg.
Rabbit bc1
Hex bipyramid P6522 212 212 354 90deg. 12 1 4.60 3.5
90deg. 120deg.
Chicken bc1
Orthorhombic P212121 176 184 242 90deg. 4 2 4.04 3.0
90deg. 90deg.
frozen- P212121 169 181 239 90deg. 4 2 3.76 2.9
90deg. 90deg.
1. AU = asymmetric unit. Mono = monomer. Dmax = d-spacing of recorded indexable
reflection with highest resolution.
2. The monoclinic beef crystals were mistakenly reported as centered orthorhombic
(C2221) in ref. 13
Distance (Å) from Fe2S2 Designation:
cluster to: (proximal/dist
al
Crystal heme bL heme c1 From heme bL )
beef P4122 (from ref 13) 27.0 31.0 proximal
chicken P212121 (+stigmatellin) 26.4 31.6 proximal
chicken P212121 34.3 21.3 distal
beef P6522 34.9 17.2 distal
beef P21 35.1 17.5 distal
rabbit P6522 35.5 19.1 distal
Iron
peaks located as peaks in electron density calculated from averaged
experimental phases improved and extended by molecular averaging, except for
chicken P212121 in the absence of inhibitor, in which case Bivoet difference
amplitudes were used with improved experimental phases retarded by 90deg..
Unique Complete-nes X-Ray1
s (%)
dmin No. Refln Refl (I > 1 Rmerge Source
sigma) (%)
Native:
chn21 3.60 279,119 70,363 76.3 (58.1) 18.6 RA
chc01 3.10 556,456 123,869 91.6 (80.5) 10.2 SSRL
chm 3.01 569,255 141,427 96.6 (74.2) 16.2 SSRL
chb 2.95 433,902 131,641 81.7 (51.2) 27.8 BNL
Derivative:
PIP2 3.50 292,339 86,221 91.8 (76.3) 10.8 SSRL
NSDMA3 3.90 425,028 67,109 99.7 (79.0) 12.4 RA
TML024 3.50 203,105 61,380 65.2 (53.5) 17.1 RA
TML034 4.30 110,201 38,103 74.4 (50.6) 21.6 RA
HPDL5 4.00 129,187 48,715 78.4 (54.0) 20.2 RA
Iridium6 3.50 177,303 68,522 71.7 (43.5) 13.4 RA
TMLssrl4 3.50 160,826 61,128 65.6 (43.9) 19.9 SSRL
HPDLssrl5 3.15 350,204 92,367 71.6 (60.2) 19.3 SSRL
1. X-ray sources and wavelengths are indicated by: RA- rotating anode
(1.54 Å). SSRL- Stanford Synchrotron Radiation Laboratory BL7-1 (1.08 Å).
BNL- Brookhaven National Laboratory X-12b (1.006 Å).
Derivatives Rderi No. of Rc Phasing power in Resolution shell (Å)
(%) sites 12.56 8.77 6.74 5.47 4.61 3.98 3.50 Total
PIP2 13.2 16 0.83 0.88 1.01 1.12 1.12 0.94 0.97 0.86 0.97
NSDMA3 15.2 16 0.88 0.68 0.61 0.76 0.96 0.93 0.96 0.97 0.89
TML024 21.5 23 0.79 1.92 1.81 1.94 1.76 1.24 1.05 0.99 1.32
TML034 28.1 23 0.71 1.84 1.67 1.88 1.67 1.38 - - 1.66
HPDL5 15.3 4 0.89 1.04 0.94 0.98 0.96 0.70 0.64 - 0.82
Iridium6 20.6 11 0.93 0.49 0.50 0.66 0.84 0.72 0.64 0.66 0.67
Figure of Merit 0.25 0.58 0.62 0.56 0.48 0.41 0.29 0.46
2. Ethylenediamine platinum iodide)2
3. N- (5-Nitrosalicyl)- (S-decylmercuri)6-aminothiophenol, a putative antimycin analog
4. Trimethyl lead acetate (different concentrations and soaking times)
5. Hexaphenyl di-lead
6. Iridium carbonyl
Figure
Legends . (b) stigmatellin binding site, density
contoured at 0.8 (blue) and 5.0 (orange) for the ironsulfur cluster. The
backbone of cytochrome b is in red and that of the ironsulfur protein is in
magenta.
. (b) stigmatellin binding site, density
contoured at 0.8 (blue) and 5.0 (orange) for the ironsulfur cluster. The
backbone of cytochrome b is in red and that of the ironsulfur protein is in
magenta.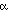 1,
1, 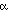 3, and
3, and 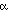 5 correspond to similarly
labeled helices in cytochrome c, while those labeled
5 correspond to similarly
labeled helices in cytochrome c, while those labeled 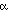 2* and
2* and
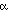 6* have no counterpart in cytochrome c.
6* have no counterpart in cytochrome c.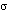 (blue) and 5
(blue) and 5 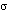 (orange). (c) contact of the Rieske protein (in the heme b distal conformation)
with cytochrome b. The model of cytochrome b (red C
(orange). (c) contact of the Rieske protein (in the heme b distal conformation)
with cytochrome b. The model of cytochrome b (red C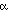 backbone plus
ball-and-stick models for residues W142 and L263 through P266) is from our
coordinates. The maps are contoured as in a. (d) interface between
cytochrome c1 and the Rieske protein. The electron density map is from
merged data of two beef hexagonal crystals, extending to 4.5 Å, phased
using experimental phases improved by density modification. The fill-in
procedure was omitted in calculating the map, which was contoured at 2
backbone plus
ball-and-stick models for residues W142 and L263 through P266) is from our
coordinates. The maps are contoured as in a. (d) interface between
cytochrome c1 and the Rieske protein. The electron density map is from
merged data of two beef hexagonal crystals, extending to 4.5 Å, phased
using experimental phases improved by density modification. The fill-in
procedure was omitted in calculating the map, which was contoured at 2
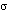 (blue) and 5
(blue) and 5 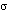 (orange). The cytochrome c1 model at right is
from our chicken bc1 model, positioned with the same operator used to position
the main part of the bc1 complex in these beef crystals.
(orange). The cytochrome c1 model at right is
from our chicken bc1 model, positioned with the same operator used to position
the main part of the bc1 complex in these beef crystals.